The Gabriella Miller Kids First Data Resource Portal houses genomic and associated clinical and phenotypic data from a variety of childhood cancer and structural birth defects cohorts. A critical next step to uncovering the causes and shared pathways underlying these conditions relies on functional validation using model systems and related tools and databases.
As a follow-up to the first two Innovation across the Phenotypic Translational Divide Webinars, the Monarch Initiative and Kids First Data Resource Center members co-hosted a third webinar on January 27, 2021 with a focus on cross-species phenotyping.
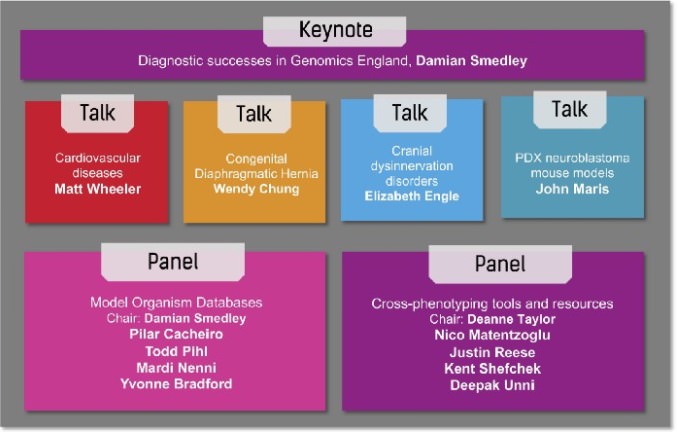
The Part 3 webinar further highlighted use cases from three Kids First X01 investigators and one other researcher, and included a keynote by Damian Smedley on diagnostic successes in Genomics England, an initiative from the UK National Health Service that aims to perform genome sequencing of cancer patients, and rare disease patients and families. In addition, there were two panel discussions led by model organism database curators and tool developers (see figure below).
Model organisms are widely used in basic science research as models for human disease, based on their similarity in anatomy, genome and phenotypes and greater accessibility for experimentation. Model Organism Databases (MODs) are valuable resources that house expert-curated data from the primary literature on organism-specific research and make it freely available to the research, clinical and patient community, often along with tools to help researchers search, analyze and visualize the data. Some example MODs include FlyBase (Drosophila), The Mouse Genome Database (mice), Saccharomyces Genome Database (baker’s yeast), and WormBase (C. elegans).
Damian Smedley from the International Mouse Phenotyping Consortium (IMPC) chaired a panel discussion featuring members from the MOD community: Pilar Cacheiro (KOMP2/IMPC — Mouse models), Todd Pihl (Integrated Canine Data Commons — Canine models), Mardi Nenni (Xenbase — Frog models) and Yvonne Bradford (ZFIN — Zebrafish models). The panelists gave some examples of the ways model organism data informed a clinical diagnosis or resulted in a high impact genetic discovery, and discussed how the data could be made even more useful to the research and medical community.
A number of tools are available to aid in query and visualization of this plethora of model organism data. Deanne Taylor from the Kids First Data Research Center presented on Integrating cross-species resources in a Common Fund Data Ecosystem to inform Kids First genetic discovery, then chaired a second panel session with panelists from the Monarch Initiative, focused on some available tools and resources for cross-phenotyping. Nico Matentzoglu discussed uPheno, the unified cross-species phenotype ontology, and the use of ontology design patterns to promote interoperability with other ontologies. Justin Reese, Kent Shefchek and Deepak Unni shared insights about the Monarch knowledge graph and Biolink API for querying and analytics. These tools are open and freely available and aim to assist with clinical diagnoses.
The brainstorming discussion focused on gathering information from the researchers about the model organisms that are best suited for studying structural birth defects and/or childhood cancer, which MODs and tools are currently in use by investigators, and barriers to performing cross-species research. This highlighted the value of chick embryo, xenopus, and zebrafish models in these fields, the importance of data standards such as ontologies, and the need to foster more collaborations.
The webinar aimed to promote new collaborations among childhood cancer and structural birth defect researchers and the MOD curators and tool developers, to build connections across resources to promote cross-species discovery with the ultimate goal of improving preventative measures, diagnostics, and therapeutics for children and families impacted by childhood cancer and structural birth defects.
To learn more about this initiative, please contact Nicole Vasilevsky (nicole@tislab.org) or Valerie Cotton (valerie.cotton@nih.gov). For questions about funding opportunities, please contact Valerie Cotton (valerie.cotton@nih.gov).
More information:
Innovation across the Phenotypic Translational Divide Webinar Part 3: Cross Species Phenotyping
When: January 27, 2021
Organizers: Nicole Vasilevsky, Valerie Cotton, Colin Fletcher, Deanne Taylor, Damian Smedley, and Melissa Haendel
Agenda: Available here
Recording: Available here
Funding: This webinar was supported by an NIH BD2K conference grant “Forums for Integrative Phenomics” to Melissa Haendel and Peter Robinson: U13CA221044.