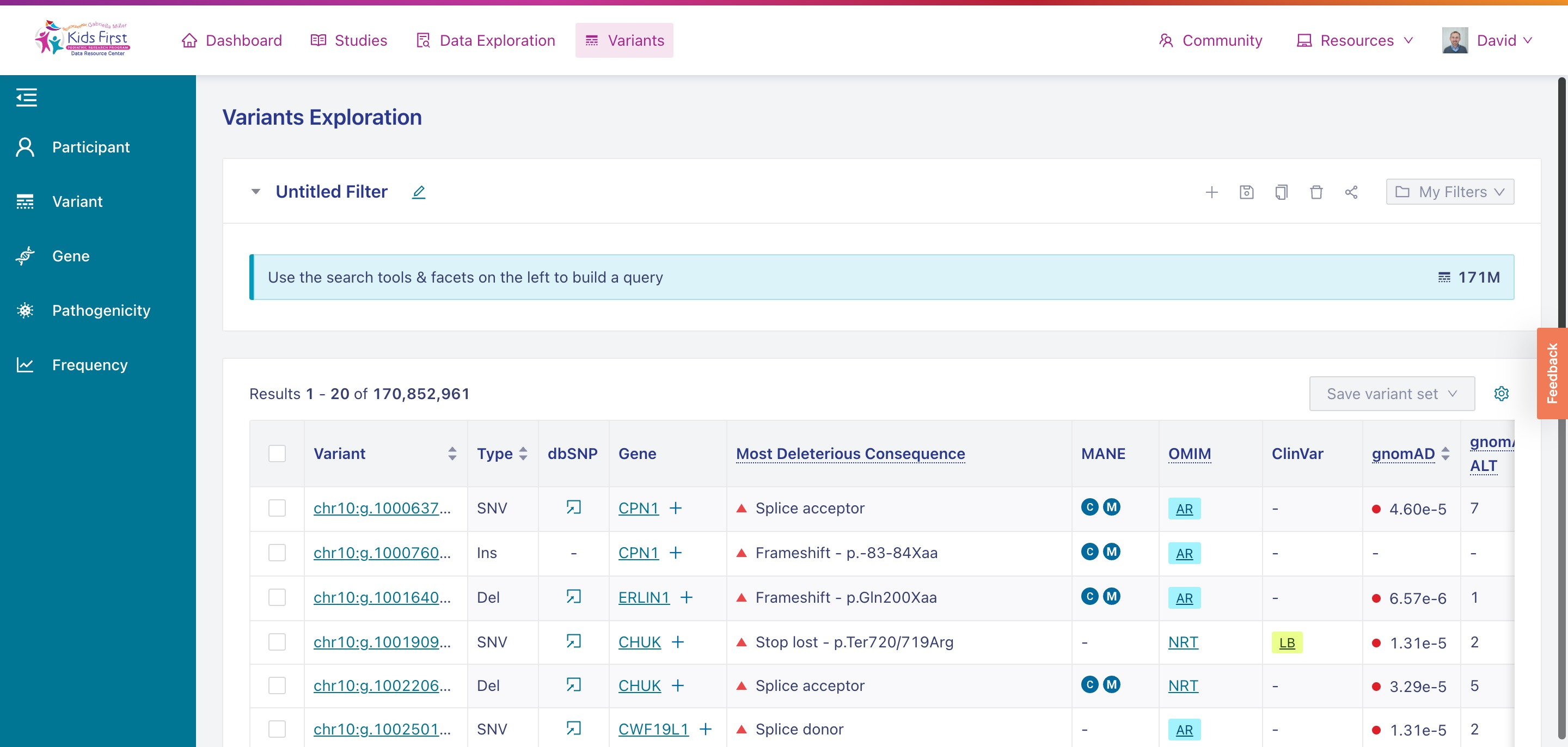
Just as Data Exploration Tool allows users to search Kids First datasets based on criteria related to participants, biospecimens, and data files, the Variant Search Tool allows users to search for germline variants present within Kids First sequenced cohorts.
Access Variant Search by clicking the Variants button in the navigation menu at the top of the screen.
The Variant Search Tool on the Kids First Portal.
Filters
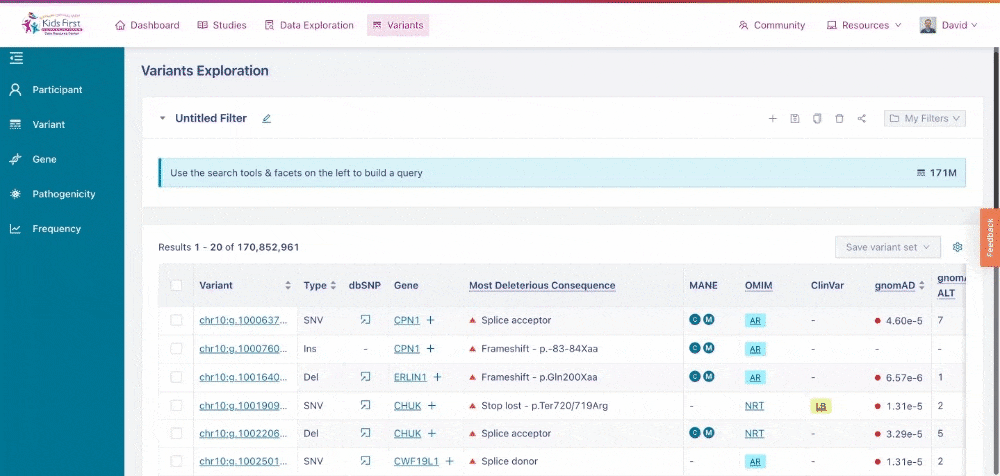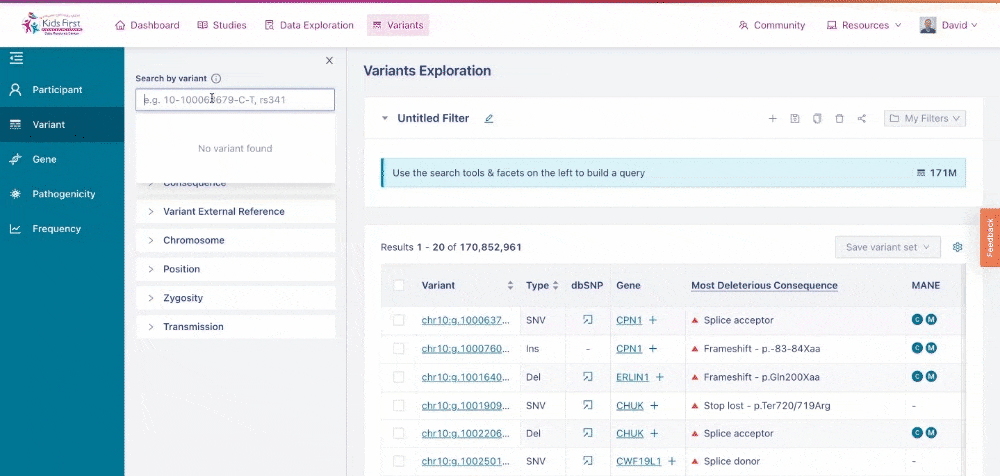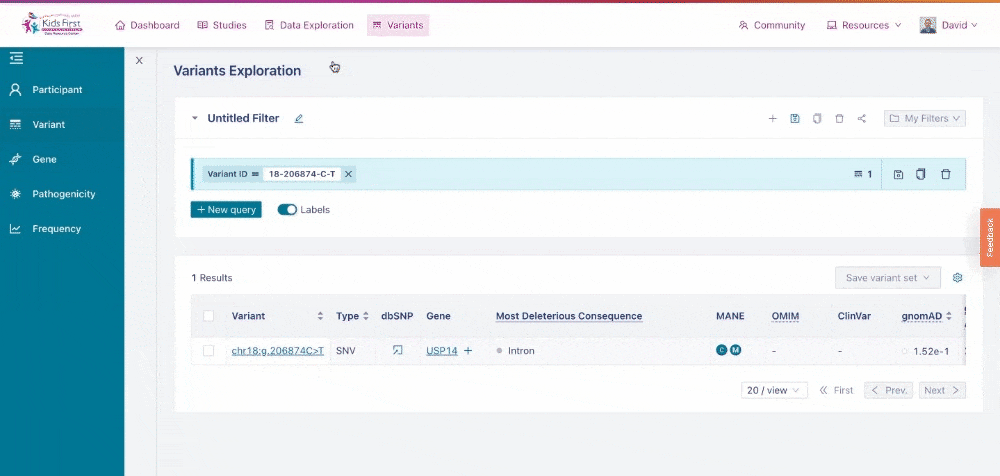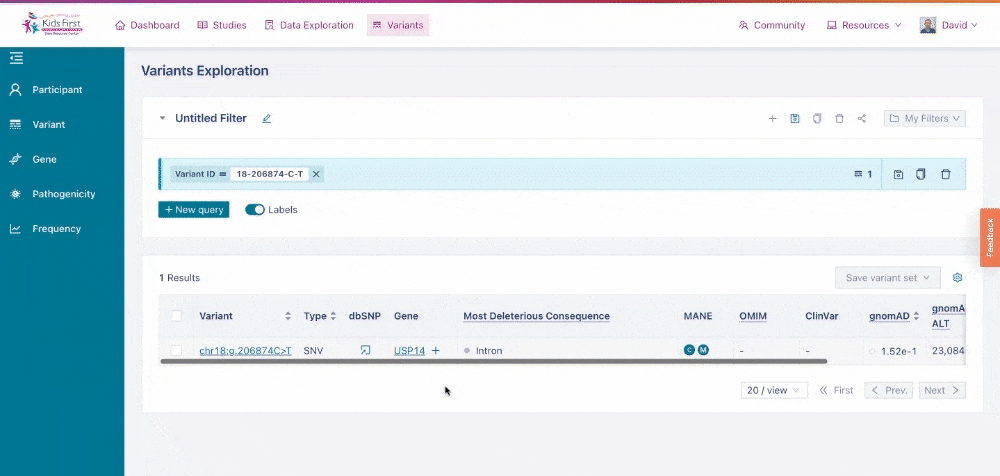
Similar to the Data Exploration Tool, users can apply filters on the collection of variants to narrow their search. Users could apply a filter to look up a single variant by dbSNP ID, for example rs572981. Choose the Variant button in the filter tab and type the ID to apply this filter.
Applying a filter for a specific variant of interest.>
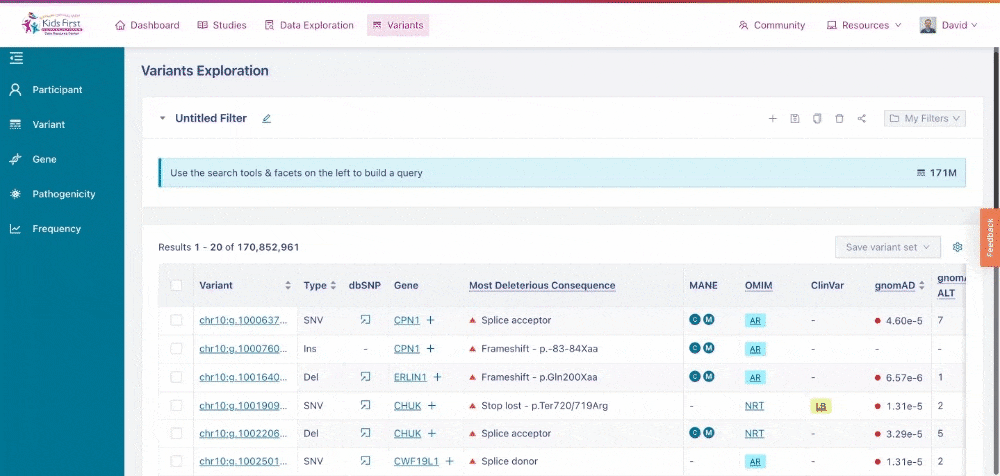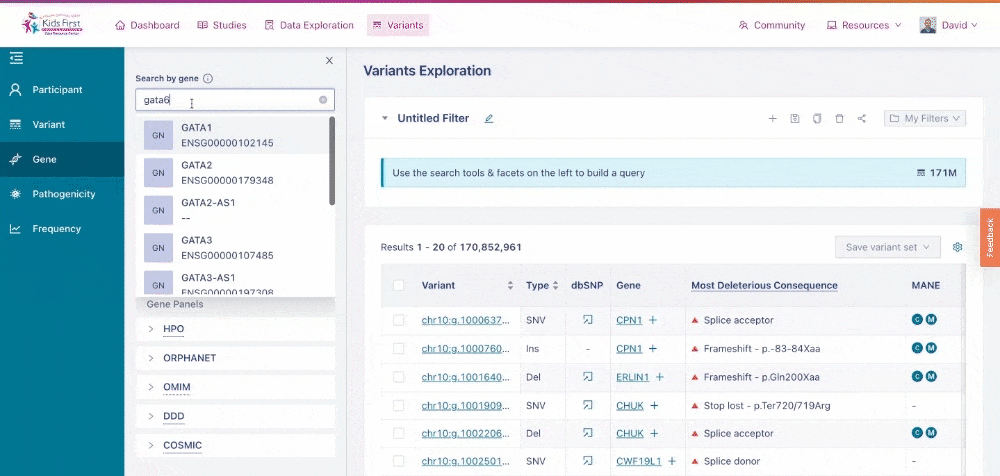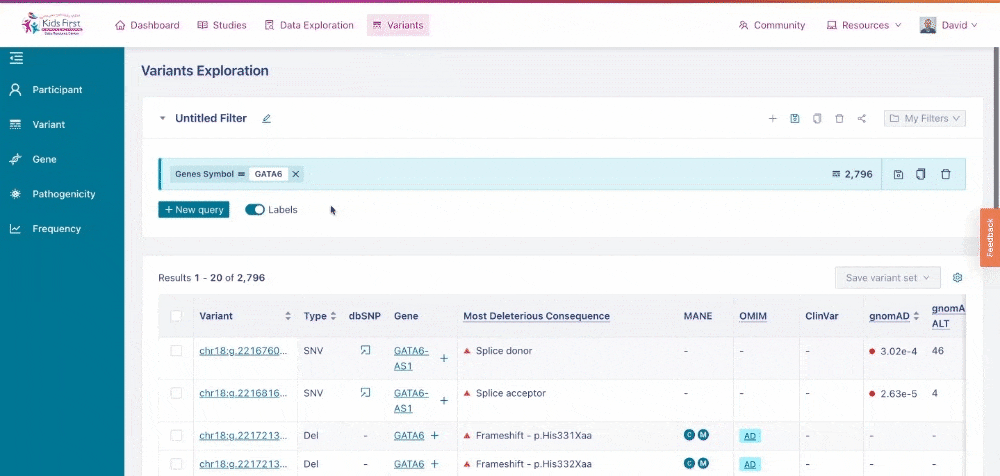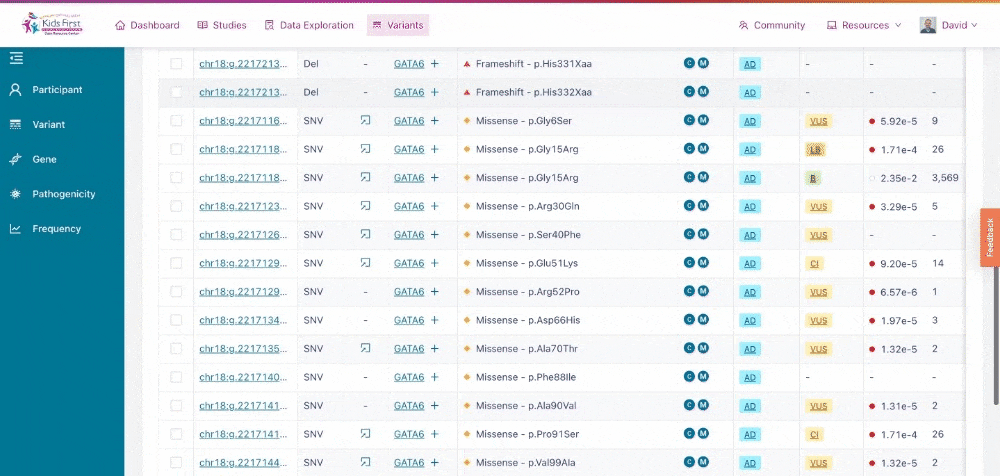
Users could also apply filters to identify variants in their gene of interest, for example GATA6. Click New query to open a new query, and then choose the Gene button in the filter tab and type the gene name to apply this filter.
Applying a filter for all variants within a specific gene of interest. Note that by clicking New query first, this opened a separate filter, rather than combining one together.
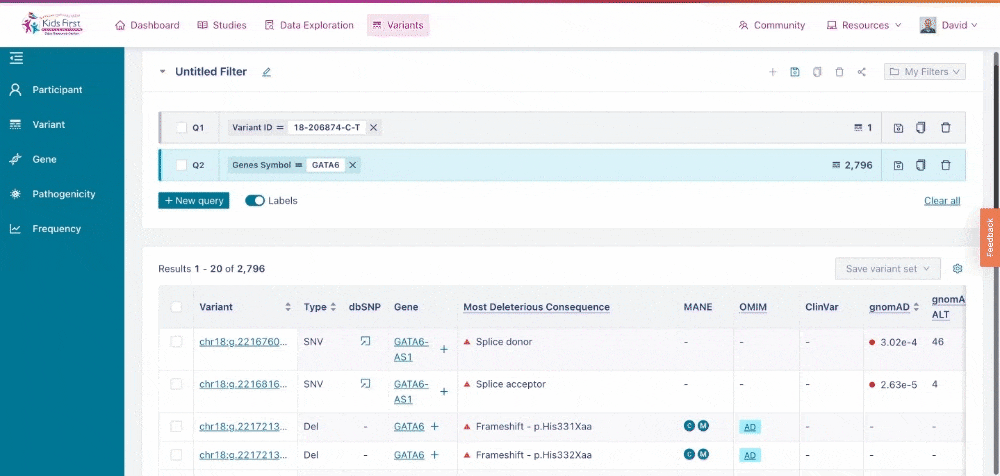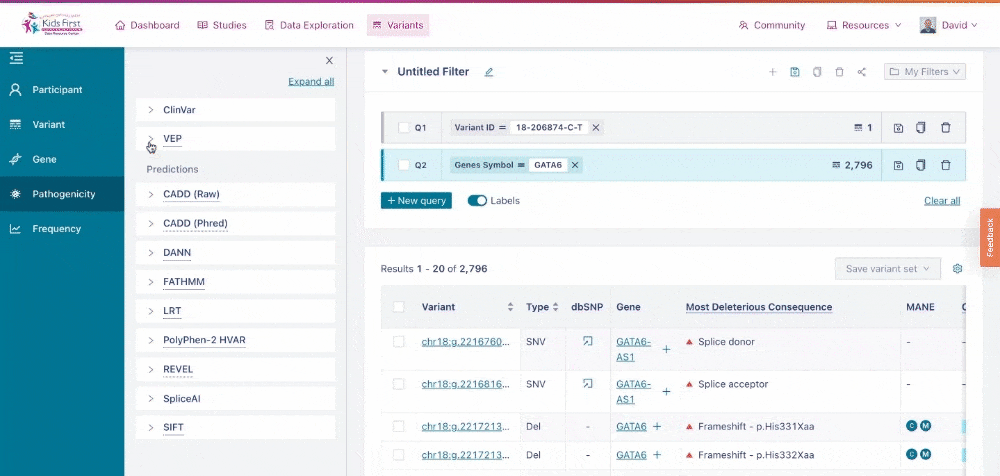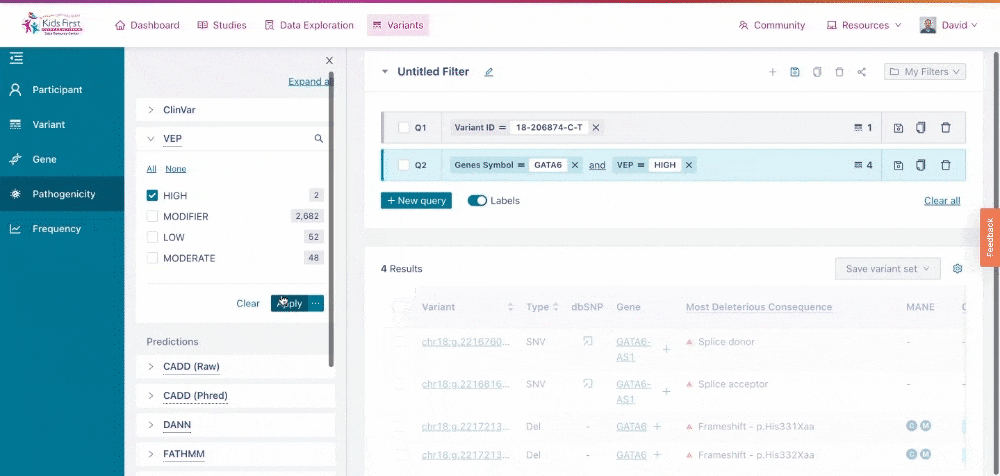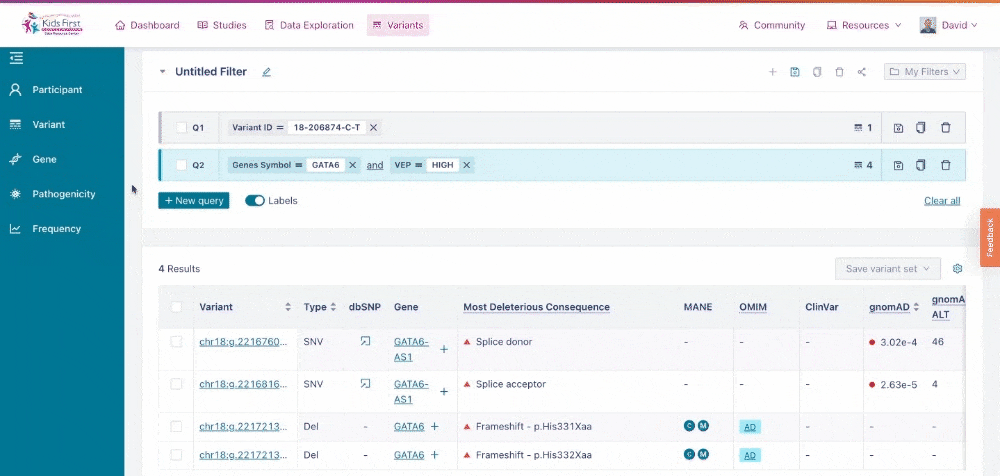
Users can narrow their search by applying filters for pathogenicity of the variants, for example just variants that scored HIGH likelihood for altering protein function according to VEP. This filter can be applied all on its own, by clicking the New query button first, or be added to the existing GATA6 query for a combined filter.
Applying a filter for pathogenicity. Because the user did not click New query, their filter was combined with the open query for variants in GATA6
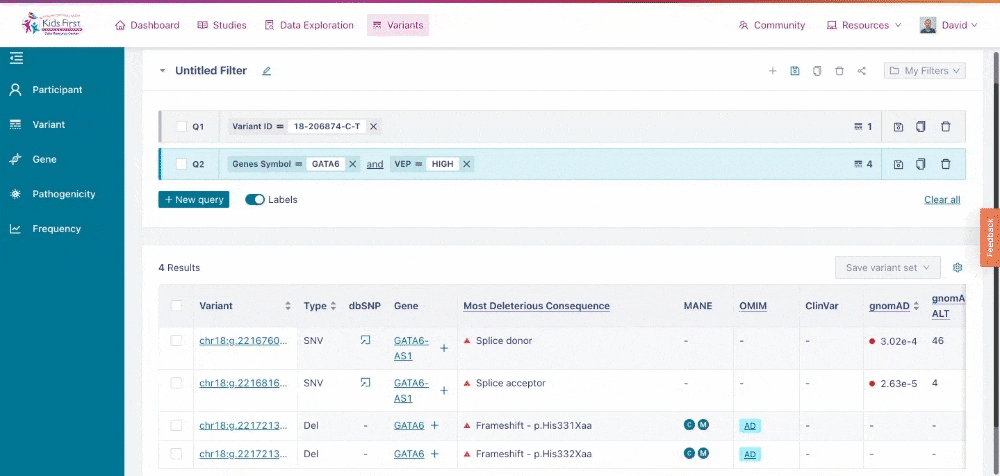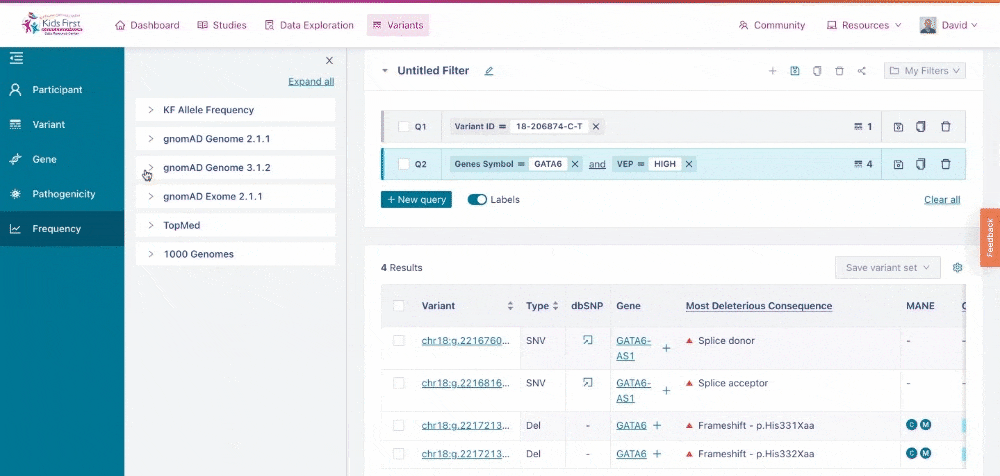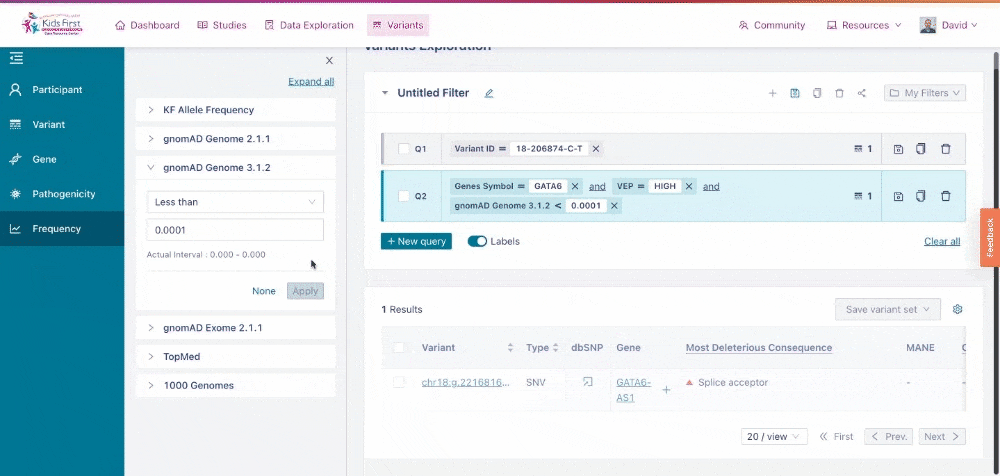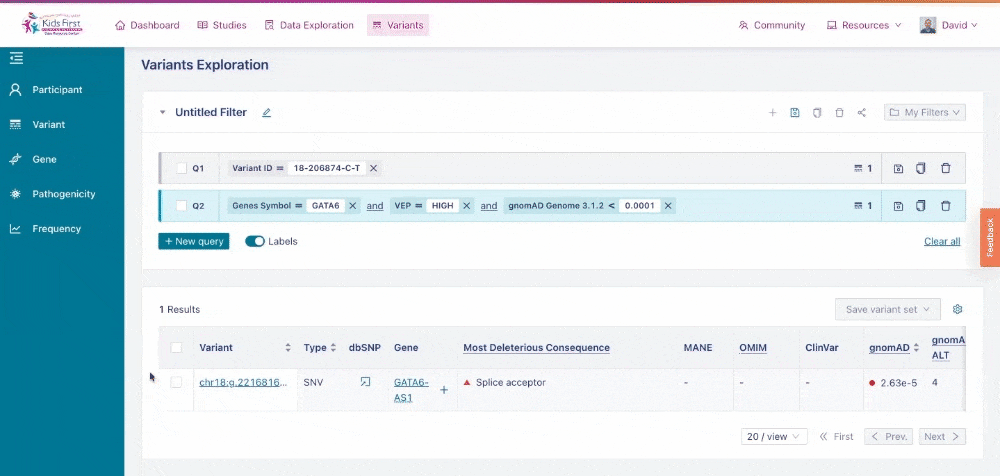
Finally, users can narrow their search results based on allele frequency of the given variants, using information from databases such as gnomAD.
Applying a filter for allele frequency. Again, because the user did not click New query, their filter was combined with the open query for high effect variants in GATA6.
The default values for the table in the Variant Search Tool are as follows.
| Column Title | Column Description |
| Variant | The genomic coordinates of the given variant. Clicking on the coordinates opens the Variant Page (see below). |
| Type | The type of genomic alteration the variant is |
| dbSNP | Links out to the NIH dbSNP database for the given variant, if available |
| Gene | The gene name associated with the most deleterious consequence the variant causes. Clicking the gene name links out to the OMIM database for the given gene. Clicking the + symbol applies a filter for just variants within that gene. |
| Most Deleterious Consequences | The most deleterious functional consequences of the given variant, as annotated using VEP |
| MANE | Annotation provided by the Matched Annotation from NCBI and EMBL-EBI project |
| OMIM | The mode of inheritance for the variant of interest |
| ClinVar | Annotation on the pathogenicity of the variant of interest from the Clinvar database |
| gnomAD | Allele frequency of the variant of interest in gnomAD Genome 3.1.2 |
| gnomAD ALT | Alternate allele count for the variant of interest in gnomAD Genome 3.1.2 |
| Part. | The number (and percentage) of Kids First participants which have the variant of interest. If there is a sufficient number of participants, clicking this link will open a query for them in the Data Exploration Tool. |
| Studies | The number of Kids First studies which have participants which have the variant of interest. Clicking this link will open a query for these studies in the Data Exploration Tool. |
Variant Page
Clicking the coordinates of a variant opens the Variant Page for that given variant.
Opening the Variant Page from within Variant Search.
The Variant Page contains more detailed information about the variant of interest than is displayed on the table view in Variant Search.
Summary: provides summary information about the variant, similar to what is shown on the Variant Search main page
Consequence: provides detailed information about the consequences of the variant of interest on each gene and transcript it affects
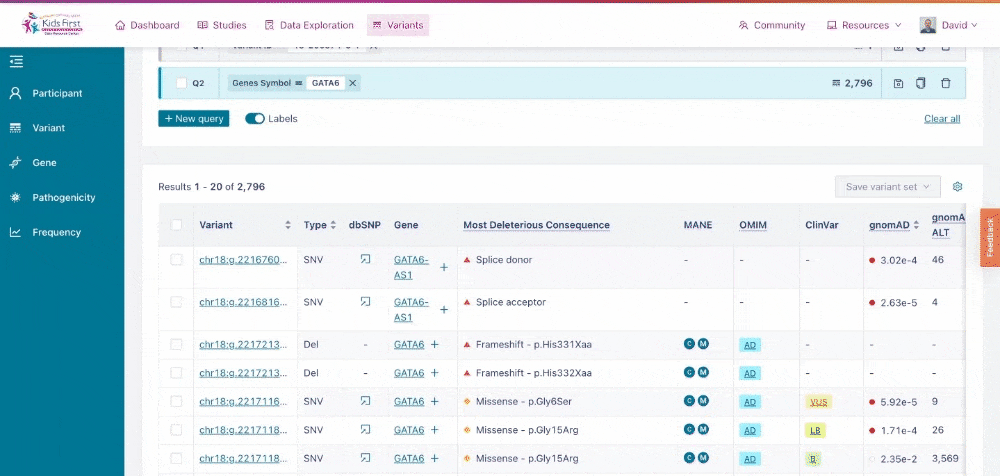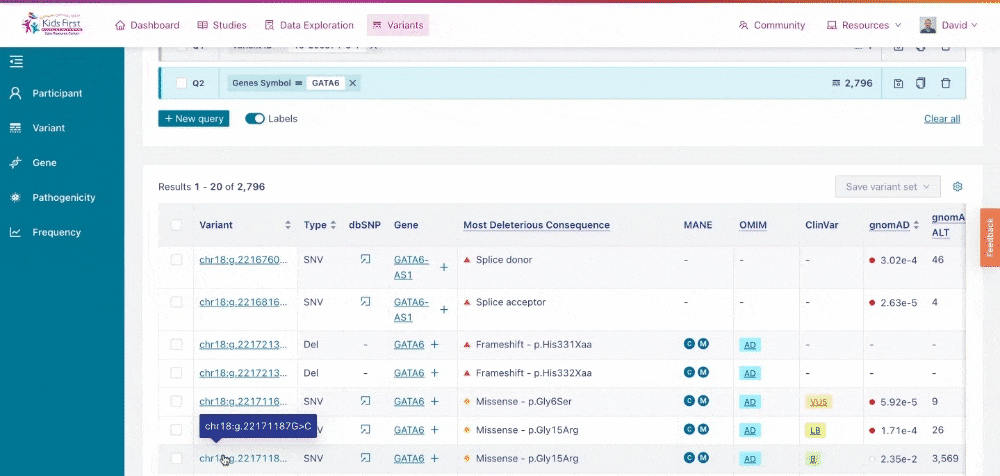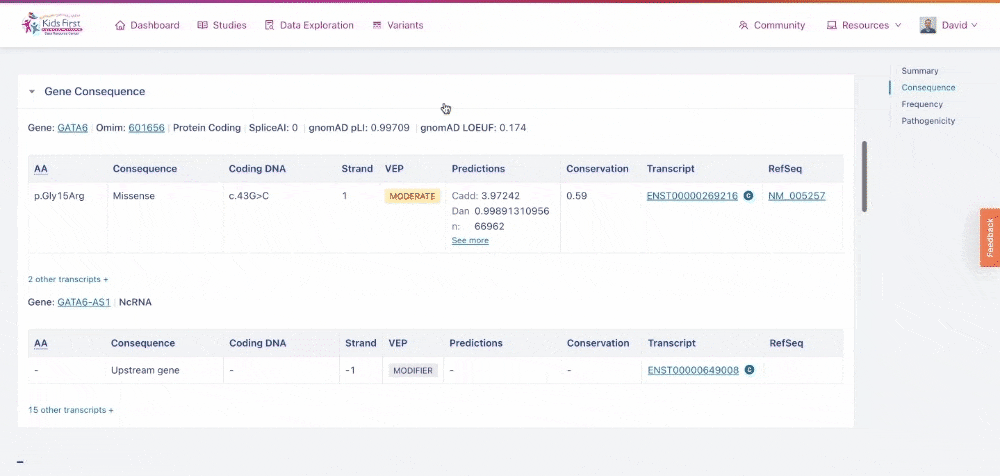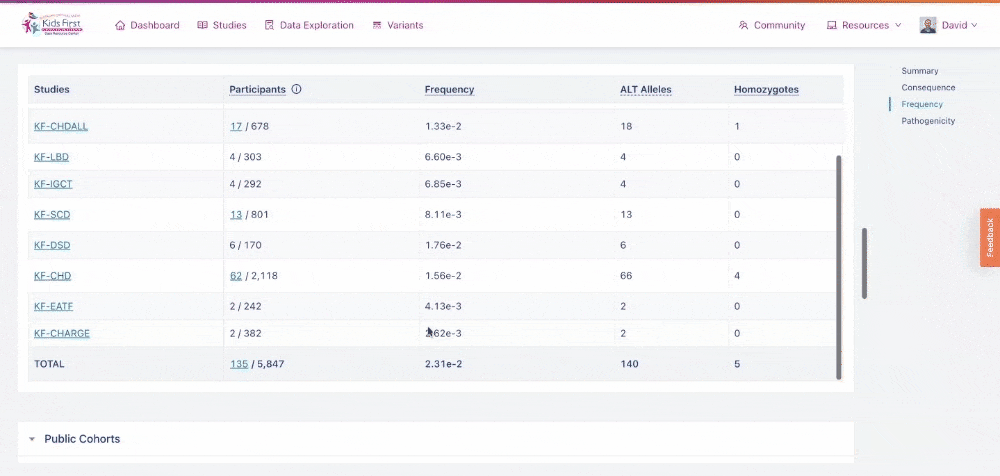
Frequency: provides information about the frequency of the variant in Kids First studies, as well as publicly available cohorts
- The Kids First table provides information about which Kids First studies the participants with the variant are enrolled in. For common variants, where there are more than 10 individuals in a study with the variant of interest, the number of participants is linkable and opens a Data Exploration query with those participant IDs loaded.
The Kids First allele frequency of rs116262672, as shown in the Variant Page. Note that participants in some studies are linkable to a Data Exploration query while others are not.
Pathogenicity: provides information about known links between the variant of interest and human disease, through databases such as ClinVar, OMIM and HPO